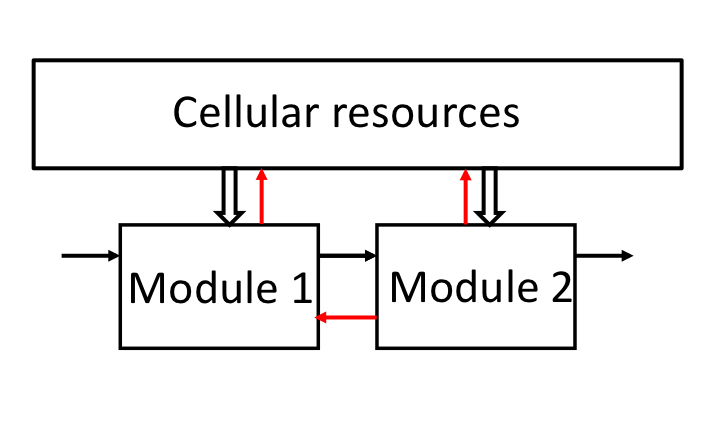
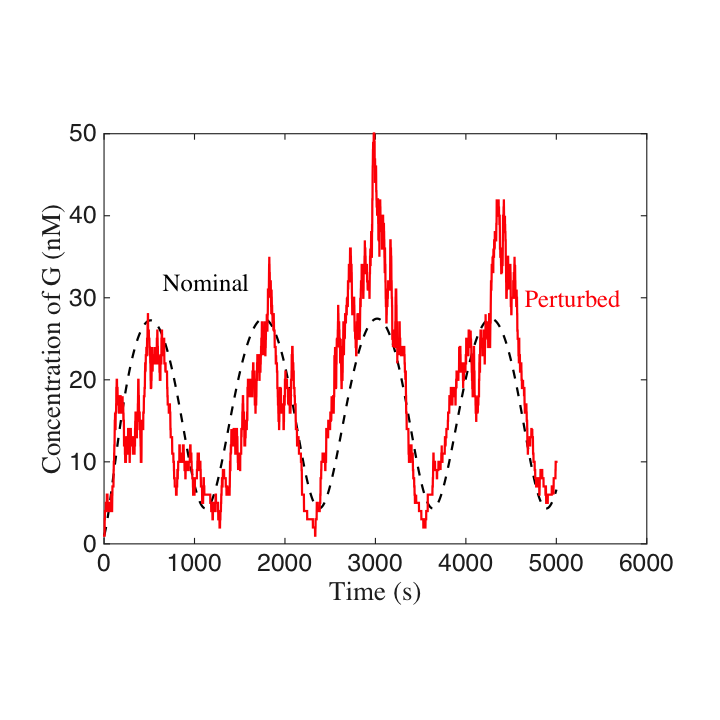
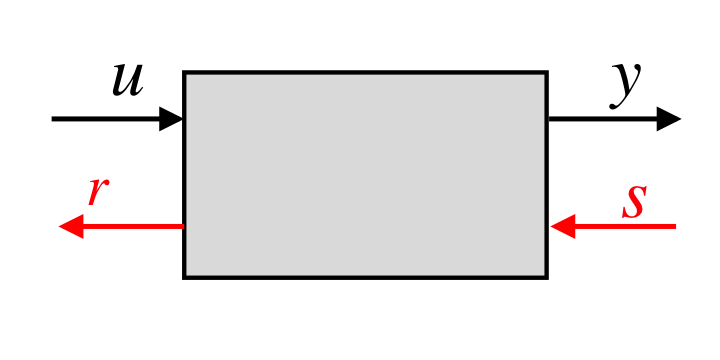
 Our system concept models load effects as retroactivity signals. Based on this concept, we tackled the load problem in synthetic biology employing disturbance attenuation techniques from control theory
Our system concept models load effects as retroactivity signals. Based on this concept, we tackled the load problem in synthetic biology employing disturbance attenuation techniques from control theory
Related Publications:
Link 1
Link 2
Link 3
Link 4
Link 5
Link 6
Link 7
Link 8
Link 9
Link 10
Link 11
Link 12
Link 13
Research
Modular design of biological networks
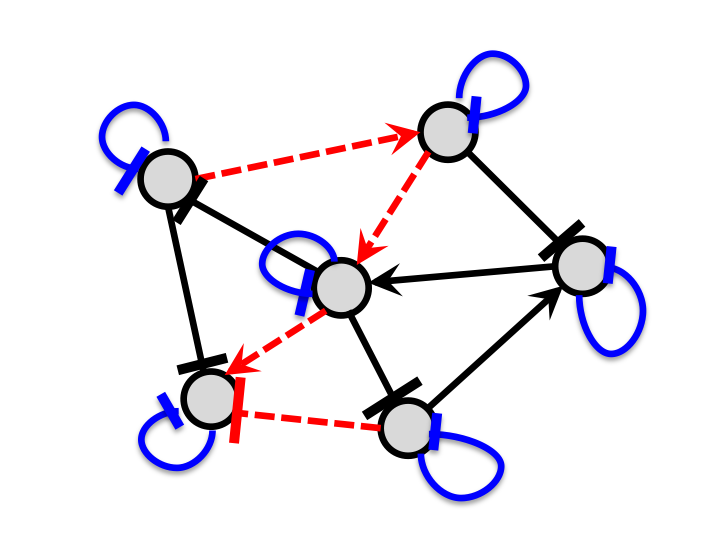
Our current ability of designing synthetic genetic circuits that behave as predicted is severely limited by the fact that a circuit’s behavior depends on its context. Because of context-dependence, the input/output behavior of a circuit is not modular and can change depending on the system in which it is inserted. This leads to an ad hoc, lengthy and combinatorial design process wherein a system’s components are co-optimized to work well with each other and need to be re-designed if used in a different system. Our group has been developing solutions to a few root causes that contribute to context-dependence: retroactivity, that is, loading due to direct connectivity, and indirect connectivity arising from resource sharing. In particular, we are developing a basic scientific understanding of these effects, predictive and design-oriented models, and engineering solutions. These solutions are based on formulating the problem of context-dependence as a problem of robustness to specific perturbations, leading to a natural design approach grounded on the fundamentals of feedback control theory. Due to the peculiar mathematical descriptions of the physical processes underlying biomolecular reactions, our circuit implementations of general feedback control principles often require the development of new control theoretic approaches to robustness. All our models and engineering solutions are tested in bacterial cells in our lab.

Gallery
Stochastic behavior
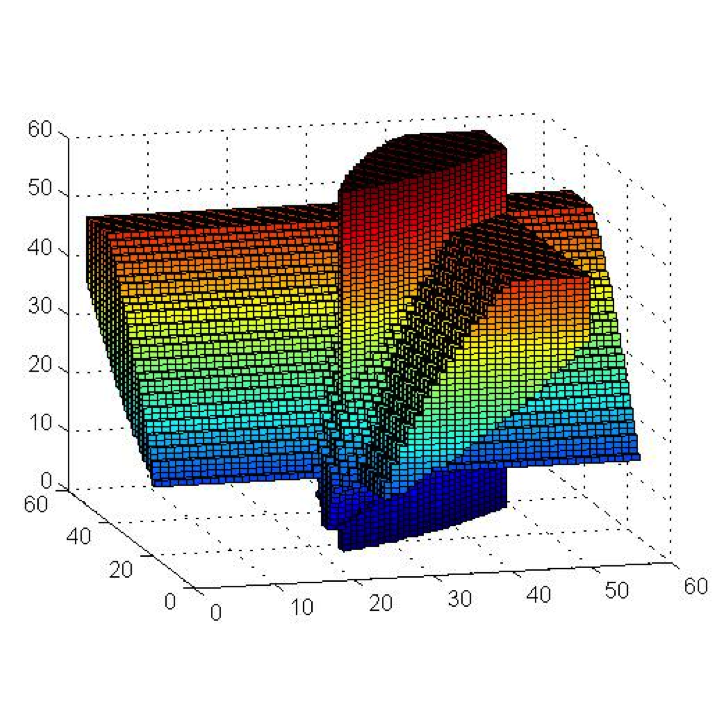
The behavior of biomolecular systems in living cells is noisy due to the intrinsic stochasticity of biochemical reactions. Stochasticity leads to subtle tradeoffs in the design of synthetic circuits and also results in stability landscapes that differ from those of the corresponding deterministic models. Time scale separation among reactions is typical in any realistic biological network and it shapes stochastic properties in fairly subtle ways. We have been developing model reduction techniques for multi-time scale stochastic systems with the main objective of deriving reduced systems that can be analytically studied. We are using these techniques to understand the relationship among multi-stability, multi-modality, and time scale separation, with application to the control of natural multi-stable gene regulatory networks.

Gallery
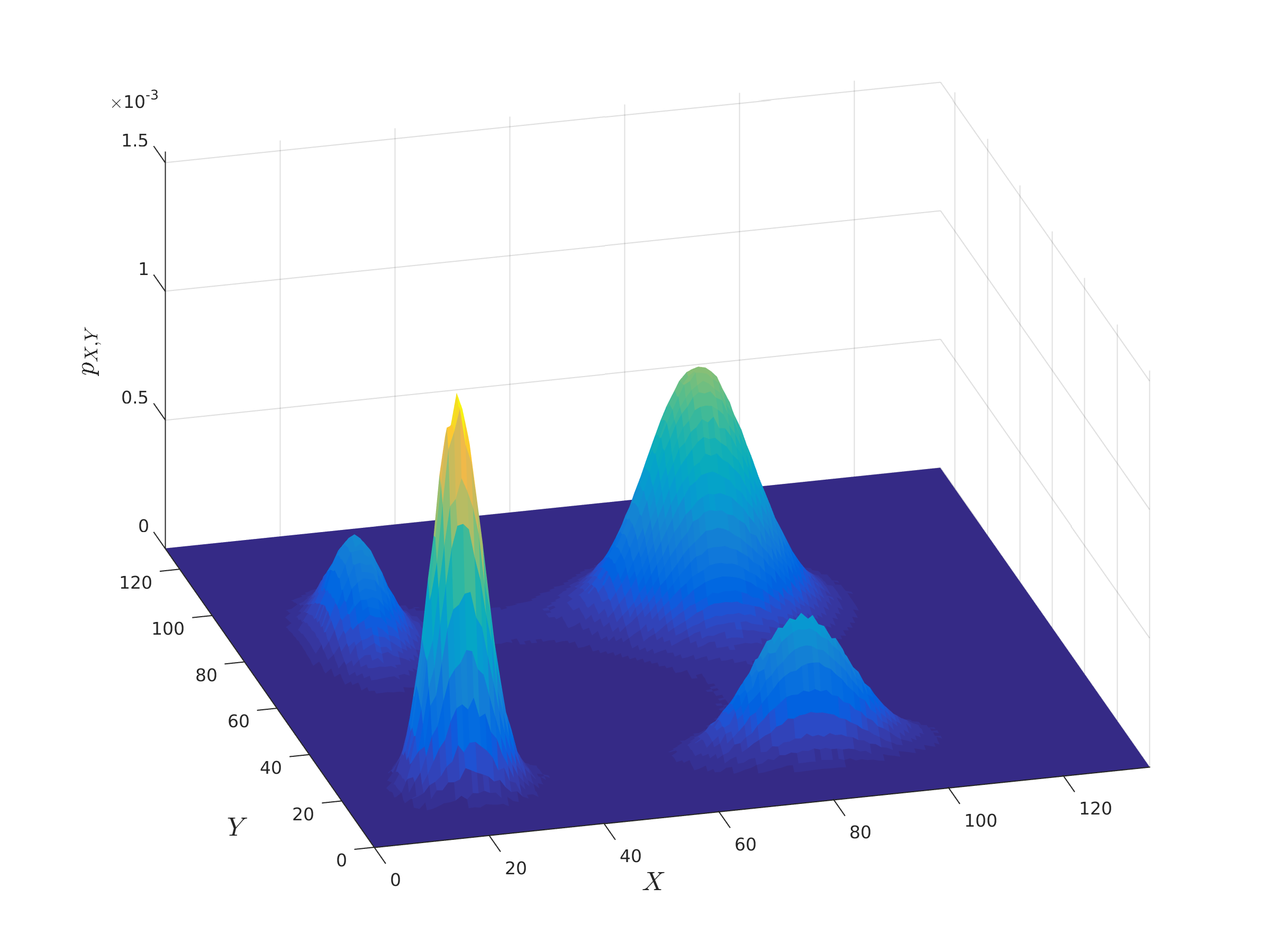 We are developing algorithms that determine when a gene regulatory network displays noise-induced multi-modality
We are developing algorithms that determine when a gene regulatory network displays noise-induced multi-modality
Related Publications:
Genetic feedback controllers to reprogram cell fate
The fate of a cell is encoded by a specific signature of transcription factor (TF) levels. Gene regulatory networks (GRNs), in turn, dictate TF levels and hence the landscape of possible cell fates. These networks determine the likelihood of transitioning among cell fates under the influence of noise or external stimulations. Controlling cell fate is therefore contingent upon the ability of accurately steering endogenous TF levels to desired values, despite the presence of a GRN that naturally regulates such levels. In this project, we are studying the implications of the dynamics of natural GRNs, including epigenetic modifications, on our ability to artificially control cell fate through artificial stimulation. Based on these studies, our objective is to design genetic feedback controllers, which can be delivered to cells through either mRNA or DNA, in order to accurately control TF levels and trigger transitions into desired cell fates. As a test-bed application, we are considering induced pluripotent stem cell (iPSC) reprogramming, and are aiming at developing a tool that can be used for any cell-fate conversion.

Gallery
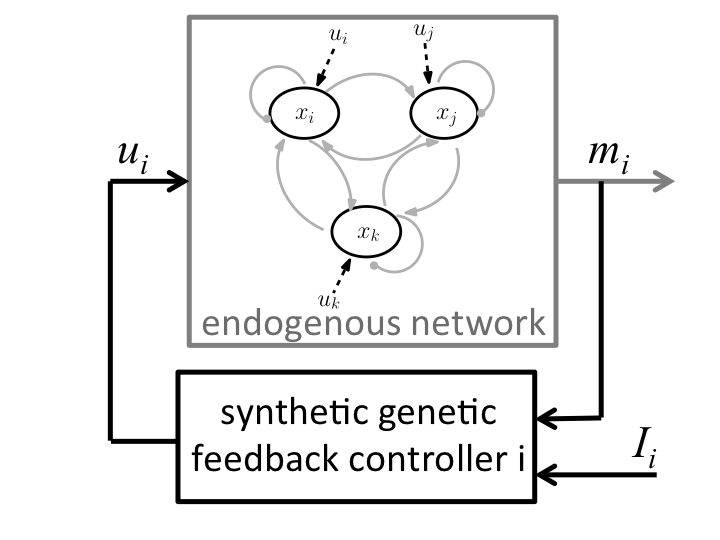 feedback control of endogenous multi-stable networks to trigger desired transitions
feedback control of endogenous multi-stable networks to trigger desired transitions
Related Publications:
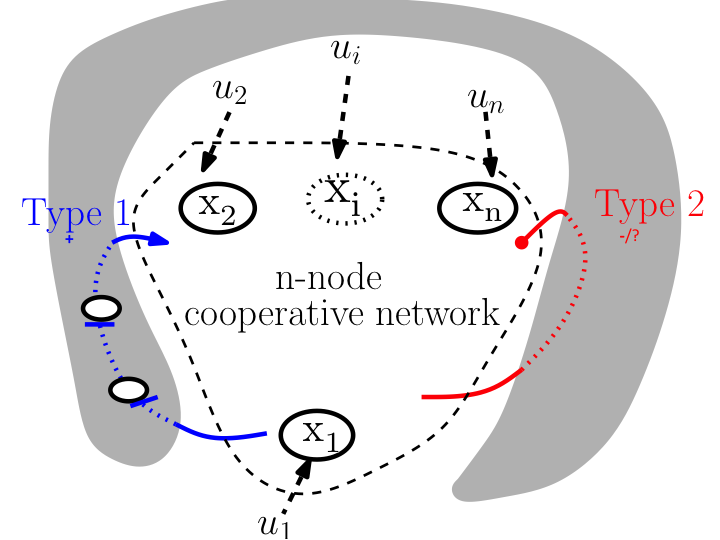 When is it possible to trigger a desired transition in a multistable gene regulatory network with experimentally realizable inputs? This question is crucial for controlling cell fate
When is it possible to trigger a desired transition in a multistable gene regulatory network with experimentally realizable inputs? This question is crucial for controlling cell fate
Related Publications:
Intelligent Transportation
(Archived- no longer active) We have been working since 2006 on (cooperative) active safety systems to prevent collisions with focus on traffic intersections. Our approach is controltheoretic and based on the computation of the capture set, the set of states from which collisions are unavoidable with our control freedom. The strategy of the driver assist system is to be silent unless the capture set is hit, at what point it is warning first and then forcing specific throttle brake combinations to avoid entering the capture set.