High Level Research
***********************************2024: LOOKING FOR NEW GRADUATE STUDENTS TO START IN SEPTEMBER********************************************* Del Vecchio’s group focuses on model-based analysis, design, and control of biomolecular circuits in living cells, both bacterial and mammalian. A core foundational problem we are interested in is context-aware design strategies for genetic circuits, such that desired behavior is conserved across different genetic and cellular contexts. Our main current applications are: multiplexed bio-sensing in bacteria, cell fate reprogramming including control of chromatin state through novel genetic circuits, and design of artificial cell fate differentiation circuits in bacteria and mammalian cells.
Research

Modular design of biological networks
Our current ability of designing synthetic genetic circuits bottom up, that is, our ability to create larger systems from the composition of simpler functional units...
Stochastic behavior
The behavior of biomolecular systems in living cells is noisy due to the intrinsic stochasticity of biochemical reactions. Stochasticity leads to subtle tradeoffs in the design of synthetic circuits...
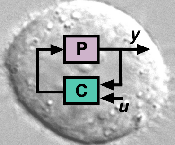
Genetic feedback controllers to reprogram cell fate
(Post-doc position available for experimental work) The fate of a cell is encoded by a specific signature of transcription factor (TF) levels. Gene regulatory networks (GRNs), in turn, dictate TF levels...

Intelligent Transportation
(Archived- no longer active) We have been working since 2006 on (cooperative) active safety systems to prevent collisions with focus on traffic intersections...
Latest Publications:
Guaranteeing System-level Properties in Genetic Circuits Subject to Context Effects
Inigo X. Incer, Ayush Pandey, Nicholas Nolan, Emma L. Peterman, Kate E. Galloway, Eduardo D. Sontag, and Domitilla Del Vecchio
Submitted
March 2024
Assessing Feasibility in Resource Limited Genetic Networks
Carlos Eduardo Celeste Junior, Theodore Wu Grunberg and Domitilla Del Vecchio
Submitted
March 2024
Competition for binding targets results in paradoxical effects for simultaneous activator and repressor action
M. Ali Al-Radhawi, Krishna Manoj, Dhruv Jatkar, Alon Duvall, Domitilla Del Vecchio, and Eduardo D. Sontag
Submitted
March 2024